Structure of DNA and RNA:
- The image below depicts different levels of DNA packaging.
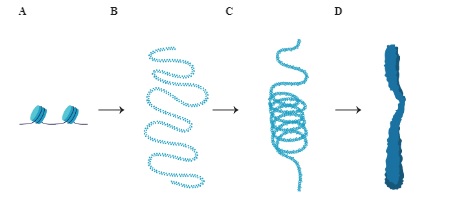
- Among the above structures, non-histone chromosomal (NHC) proteins in order to enable efficient packaging of DNA into D. If D is a chromosome, what structure does C represent?
- Chromatin fiber
- NHC proteins are required for higher order packaging of chromatin into fibers and further into chromosomes.
- Chromatin fiber
- During metaphase, structure C is coiled and condensed with the help of non-histone chromosomal (NHC) proteins into structure D. Identify C and D.
- C is a chromatin fiber, and D is a chromosome.
- Chromatin fibers are coiled and condensed into chromosomes during metaphase.
- A and B, depicts the packaging of nucleosomes into chromatin.
- Chromatin fibers are coiled and condensed into chromosomes during metaphase.
- C is a chromatin fiber, and D is a chromosome.
- Euchromatin appears as loosely-packed, lightly-stained bands within the cell when viewed under a microscope. Heterochromatin, on the other hand, is seen to be more densely packed and darkly-stained. Keeping these facts in mind, why is euchromatin transcriptionally active, while heterochromatin is not?
- Loose packing allows access to DNA for transcription.
- Loosely packed DNA in euchromatin can be accessed by transcription machinery, unlike in more densely packed heterochromatin.
- Loose packing allows access to DNA for transcription.
- The image below depicts the structural difference between euchromatin and heterochromatin.

- Which of the following statements about these structures are true?
- Heterochromatin is transcriptionally inactive.
- The dense packaging of heterochromatin makes DNA inaccessible to the transcription machinery.
- Heterochromatin is transcriptionally inactive.
- The table below contains information about the nature of certain amino acids.
| Amino acid | Charge |
| Arginine | Positive |
| Aspartic acid | Negative |
| Glutamic acid | Negative |
| Leucine | Neutral |
| Lysine | Positive |
- Which of these amino acids can be added to histones to enable binding of DNA?
- Arginine
- Histones contain positively-charged amino acids like arginine, which allow them to closely interact with and bind negatively-charged DNA.
- Lysine
- Histones contain positively-charged amino acids like lysine, which allow them to closely interact with and bind negatively-charged DNA.
- Arginine
- Amino acids such as arginine (Arg) and lysine (Lys) are commonly found in histones. Glutamic acid (Glu) and aspartic acid (Asp), on the other hand, are unlikely to be present. Which of the following statements correctly explain the reason for this difference?
- Arg and Lys are positively charged, while Glu and Asp are negatively charged.
- Basic, positively-charged residues like Arg and Lys can bind negatively-charged DNA better than negatively-charged acidic residues like Glu and Asp.
- Arg and Lys are positively charged, while Glu and Asp are negatively charged.
- Which of the following components would a nucleosome contain?
- Histone octamer + DNA
- Nucleosomes consist of about 200bp of DNA wrapped around histone octamers.
- Histone octamer + DNA
- The average human cell has about 30 million ‘bead’-like structures in its nucleus. Each ‘bead’ contains roughly 200bp of DNA wrapped around an octamer of proteins. What structure does this ‘bead’ represent?
- Nucleosome
- Histone octamers along with the associated DNA form a structure known as a nucleosome, which has a bead-like appearance when seen through a microscope.
- Nucleosome
DNA and RNA structure:
- The figure below represents a prokaryotic cell. Inside the cell, different structures containing hereditary information are shown. Which of the following statements correctly compares Structures A and B?

- Structures A and B are both made up of double-stranded DNA.
- According to the text, Structures A and B both contain hereditary information. Structure A is shown in the diagram as a large, densely-coiled mass, so it is likely a chromosome. In contrast, Structure B is shown as a small circle, so it is likely a plasmid. In prokaryotic organisms, chromosomes and plasmids are both made up of double-stranded DNA.

- The figure above represents a prokaryotic cell. Inside the cell, different structures containing hereditary information are shown.
- Which of the following statements correctly compares Structures 1 and 2?
- Structures 1 and 2 are both circular.
- According to the text, Structures 1 and 2 both contain hereditary information. Structure 1 is shown in the diagram as a large, densely-coiled mass, so it is likely a chromosome. In contrast, Structure 2 is shown as a small circle, so it is likely a plasmid. In prokaryotic organisms, chromosomes and plasmids are both circular.
- Structures 1 and 2 are both circular.
- Which of the following statements correctly compares Structures 1 and 2?
- Feline panleukopenia is a highly contagious and often fatal illness seen in both wild and domestic cats. The infection is caused by the feline panleukopenia virus, also known as FPLV.
The nucleotide composition of the FPLV genome is shown in the following table.
| Nucleotide | Percentage of FPLV genome |
| Guanine (G) | 19.9% |
| Thymine (T) | 28.2% |
| Cytosine (C) | 15.7% |
| Adenine (A) | 36.2% |
According to the data in the table, the FPLV genome is made up of
- single-stranded DNA
- Zika fever is an illness caused by the Zika virus, which is spread to people by mosquitoes of the genus Aedes.
The nucleotide composition of the Zika virus genome is shown in the following table.
| Nucleotide | Percentage of Zika virus genome |
| Adenine (A) | 27.7% |
| Uracil (U) | 21.3% |
| Cytosine (C) | 21.9% |
| Guanine (G) | 29.1% |
According to the data in the table, the Zika virus genome is made up of single-stranded RNA.
- Chicken pox is caused by the varicella-zoster virus, also known as VZV. The nucleotide composition of the VZV genome is shown in the following table.
| Nucleotide | Percentage of VZV genome |
| Adenine (A) | 27% |
| Cytosine (C) | 23% |
| Thymine (T) | 27% |
| Guanine (G) | 23% |
According to the data in the table, the VZV genome is made up of
- double-stranded DNA
- A scientist designs a primer, which is a short oligonucleotide, for use in a quantitative polymerase chain reaction (qPCR). During the reaction, the primer will bind to its DNA template via complementary base-pairing. The sequence and orientation of the primer is shown below.
- 5’-AGCTGAGATCGATCGATA-3’
- Which of the following shows the sequence and orientation of the primer’s DNA template?
- 3’-TCGACTCTAGCTAGCTAT-5’
- 5’-AGCTGAGATCGATCGATA-3’
- A scientist designs a primer, which is a short oligonucleotide, for use in a DNA sequencing reaction. The primer is complementary to the DNA template shown below. 3’-AGCTAGCGATCGGACGAT-5’ Which of the following shows the sequence and orientation of the primer?
- 5’-TCGATCGCTAGCCTGCTA-3’
- A scientist designs a primer, which is a short oligonucleotide, for use in a polymerase chain reaction (PCR). During the reaction, the primer will bind to its DNA template via complementary base-pairing. The sequence and orientation of the primer is shown below. 5’-CTTAGGTCATCAGACTAG-3’ Which of the following shows the sequence and orientation of the primer’s DNA template?
- 3’-GAATCCAGTAGTCTGATC-5’
- The following diagram shows nucleotide base-pairing in a double-stranded DNA molecule. The nitrogenous bases in two of the nucleotides are labeled. Which of the following correctly compares nitrogenous bases A and B?
- Nitrogenous base A is a purine, while nitrogenous base B is a pyrimidine.

- The following diagram shows nucleotide base-pairing in a double-stranded DNA molecule. The nitrogenous bases in two of the nucleotides are labeled. Which of the following correctly compares nitrogenous bases 1 and 2?
- Nitrogenous bases 1 and 2 are both pyrimidines.

- The following diagram shows nucleotide base-pairing in a double-stranded DNA molecule. The nitrogenous bases in two of the nucleotides are labeled. Which of the following correctly compares nitrogenous bases 1 and 2?
- Nitrogenous base 1 is a pyrimidine, while nitrogenous base 2 is a purine.

- The figure below represents a eukaryotic cell. In the nucleus of the cell, different structures containing hereditary information are shown. Which of the following statements correctly compares the structures labeled A and B?

- Structure B contains fewer genes than Structure A.
- According to the text, Structures A and B both contain hereditary information. Structure A is shown in the diagram as being long and constricted in the middle, so it is likely a chromosome. In contrast, Structure B is shown as being small and circular, so it is likely a plasmid. Due to their smaller size, plasmids typically contain fewer genes than chromosomes.
Nucleic acids:
- Which of the following strands are complementary DNA sequences?
- 5’CTGTCA 3′ and 5′ TGACAG 3′
- Remember that DNA runs from 5′ to 3′, so this is really asking if the complementary pairing looks like this:
- 5′ CTGTCA 3′ and 3′ GACAGT 5′
- Each nucleotide is appropriately paired according to the A-T/C-G pairing rules, so these are complementary strands.
- 5’CTGTCA 3′ and 5′ TGACAG 3′
- The percentage of thymine (T) in a given segment of double-stranded DNA is 36%, percent. What are the percentages of the remaining nucleotides?
- 36% A 14% C 14% G
- 100% – (36% T – 36% { A}) = ({G + C})
- 28% = {A + T})14% A and14% T
- Which of the following models most accurately represents the structure of DNA?

DNA is a double-stranded molecule in which guanine (G) pairs with cytosine (C) and thymine (T) pairs with adenine (A). These nucleotide bases make up the “steps” of the ladder, and sugar-phosphates make up the “backbone” of the ladder.
- In a sample of double-stranded DNA, 30%, percent of the nitrogenous bases are guanine (G). What percentage of the nitrogenous bases in the sample are adenine (A)?
- 20%
- 100% – (30% { G} – 30% { C}) = ({A + T})
- 40% = {A + T}
- 20%
- In a sample of double-stranded DNA, 30%, percent of the nitrogenous bases are adenine (A). What percentage of the nitrogenous bases in the sample are thymine (T)?
- 30%
- Due to complementary base pairing, the percent composition of thymine is the same as the percent composition of adenine.
- 30%
- In a sample of double-stranded DNA, 10% of the nitrogenous bases are guanine (G). What percentage of the nitrogenous bases in the sample are adenine (A)?
- 40%
- Guanine and cytosine are present in equal amounts of 10%each. This means the remaining 80% percent is equal parts adenine and thymine. 80%÷2=40%
- 40%
- In a sample of double-stranded DNA, 20% of the nitrogenous bases are guanine (G). What percentage of the nitrogenous bases in the sample are cytosine (C)?
- 20, percent
- Due to complementary base pairing, the percent composition of cytosine is the same as the percent composition of guanine.
- 20, percent
- Which of the following statements is true regarding DNA structure?
- DNA forms a double helix.
- DNA is made of two strands that wind around each other, forming the twisted ladder-like structure of a double helix.
- DNA forms a double helix.
- A DNA template strand contains the following sequence:
- 5′ AGGCTCCAG 3′ – Which complementary RNA strand can be made from this sequence?
- 5′ CUGGAGCCU 3′
- Remember that DNA and RNA run from 5′ to 3′, so this is really asking if the complementary pairing looks like this:
- 5′ AGGCTCCAG 3′ and 3′ UCCGAGGUC 5′ – Each nucleotide is appropriately paired according to the A-U/C-G pairing rules, so these are complementary strands.
- Remember that DNA and RNA run from 5′ to 3′, so this is really asking if the complementary pairing looks like this:
- 5′ CUGGAGCCU 3′
- 5′ AGGCTCCAG 3′ – Which complementary RNA strand can be made from this sequence?
- Identify the following statements as characteristics of DNA, RNA, or both.
| DNA | RNA | Both | |
| Contains deoxyribose | 1 | 0 | 0 |
| Is made of nucleotides | 0 | 0 | 1 |
| Forms a double helix | 1 | 0 | 0 |
| Uses uracil as a base | 0 | 1 | 0 |
| Includes phosphate groups | 0 | 0 | 1 |
| Nucleic acid | Nucleotide composition | Nitrogenous bases | Shape |
| DNA | Deoxyribose, phosphate group, nitrogenous base | Adenine, thymine, cytosine, guanine | Double helix |
| RNA | Ribose, phosphate group, nitrogenous base | Uracil, adenine, cytosine, guanine | Single helix |
DNA structure and replication:
- A template strand of DNA is 3’AACCGAGTGGA 5′ What is the complementary DNA strand that is created from this template during replication?
- 5′ TTGGCTCACCT 3′
- The question is asking if the following pairing is accurate
- 3′ AACCGAGTGGA 5′ and 5′ TTGGCTCACCT 3′
- Each nucleotide is appropriately paired according to the A-T/C-G pairing rules, so these are complementary strands.
- The question is asking if the following pairing is accurate
- 5′ TTGGCTCACCT 3′
- A template strand of DNA is 3′ TACCGATTGCA 5′. What is the complementary DNA strand that is created from this template during replication?
- 5′ ATGGCTAACGT 3′
- The question is asking if the following pairing is accurate
- 3′ TACCGATTGCA 5′ and 5′ ATGGCTAACGT 3′
- Each nucleotide is appropriately paired according to the A-T/C-G pairing rules, so these are complementary strands.
- The question is asking if the following pairing is accurate
- 5′ ATGGCTAACGT 3′
- A template strand of DNA is 3′ AACTGAACTTT 5′. What is the complementary DNA strand that is created from this template during replication?
- 5′ TTGACTTGAAA 3′
- 3′ AACTGAACTTT 5′ and
- 5′ TTGACTTGAAA 3′
- Each nucleotide is appropriately paired according to the A-T/C-G pairing rules, so these are complementary strands.
- 5′ TTGACTTGAAA 3′
- A template strand of DNA is 3′ GATCCAAAATGT 5′. What is the complementary DNA strand that is created from this template during replication?
- 5′ CTAGGTTTTACA 3′
- 3′ GATCCAAAATGT 5′ and 5′ CTAGGTTTTACA 3′
- Each nucleotide is appropriately paired according to the A-T/C-G pairing rules, so these are complementary strands.
- 3′ GATCCAAAATGT 5′ and 5′ CTAGGTTTTACA 3′
- 5′ CTAGGTTTTACA 3′
- Which of the following statements correctly describes each new molecule of DNA produced after DNA replicates?
- Each new molecule contains one strand from the parent molecule and one newly synthesized strand.
- The image below shows a DNA double helix that is about to undergo replication to produce two new DNA molecules. Which image best represents the two new DNA molecules?


Replication:
- Which of the following diagrams best represents leading strand and Okazaki fragment synthesis at a replication fork?

- Which of the following diagrams best represents the synthesis of leading strands and Okazaki fragments in a replication bubble?

- Which of the following models most accurately represents the process of DNA replication over two cell divisions?


- In which of the following diagrams of DNA replication are the enzymes correctly labeled?

- The following symbols represent enzymes involved in DNA replication.

- Given the symbols above, which of the following diagrams most accurately depicts the process of DNA replication ?

- In DNA replication, which of the following events happens during leading strand synthesis but not during lagging strand synthesis?
- DNA polymerase synthesizes a single, continuous strand of DNA.
- DNA polymerase synthesizes a single, continuous strand of DNA only during leading strand synthesis. During lagging strand synthesis, the enzyme instead synthesizes multiple short strands of DNA (i.e., Okazaki fragments).
- DNA polymerase synthesizes a single, continuous strand of DNA.
- In DNA replication, which of the following events happens during both leading and lagging strand synthesis?
- RNA primers help initiate DNA synthesis.
- RNA primers are required for DNA polymerases to begin synthesizing new strands of DNA. This is true during both leading and lagging strand synthesis.
- RNA primers help initiate DNA synthesis.
- Which of the following statements is true regarding DNA replication?
- During lagging strand synthesis, ligase joins together multiple Okazaki fragments.
- During DNA replication, multiple Okazaki fragments are joined together by ligase to form the lagging strand.
- During lagging strand synthesis, ligase joins together multiple Okazaki fragments.
Central dogma:
- Which of the following statements is true regarding transcription and translation?
- During transcription, an mRNA molecule is created from the DNA molecule.
- Transcription copies a segment of DNA into a complementary mRNA strand so that it be translated.
- During transcription, an mRNA molecule is created from the DNA molecule.
- Many antibiotics interfere with the transfer of genetic information from RNA to protein, preventing bacteria from growing. Which of the following processes is affected by antibiotics?
- Translation
- During translation, RNA is used as a code to create proteins. If antibiotics interfere with the ability for RNA to transfer information to proteins, then translation is being affected.
- Translation
- Which direction does genetic information flow during gene expression?
- DNA ⟶RNA⟶ Protein
- The central dogma of molecular biology states that DNA is transcribed to mRNA, which is translated to protein.
- The central dogma states that DNA determines an organism’s traits, such as eye color or hair color. Which sequence best represents the relationship between DNA and the traits of an organism?
- A DNA base sequence in an organism is transcribed to mRNA, which are then translated into amino acid sequences. These sequences determine protein shape, and the protein shape determines protein function. The function of proteins affects the expression of a particular trait.

- The one gene, one enzyme hypothesis is the idea that each gene encodes a single enzyme. The hypothesis has been updated based on modern discoveries. Which of the following is NOT a reason why the initial hypothesis was updated?
- Genes code only for RNA molecules, not for enzymes.
- Genes do code for enzymes and other proteins, but they can also code for functional RNA as well.
- Genes code only for RNA molecules, not for enzymes.
- Which of the following statements is true regarding transcription and translation?
- During transcription, an mRNA molecule is created from the DNA molecule.
- Transcription copies a segment of DNA into a complementary mRNA strand so that it be translated.
- During transcription, an mRNA molecule is created from the DNA molecule.
Transcription:
- The following RNA strand was produced:
- 5′ AAA AUG AGU AAG 3′. Which of the following DNA strands could have been the template for this RNA?
- 3′ TTT TAC TCA TTC 5′
- The question is asking if the following pairing is accurate
- RNA – 5′ AAA AUG AGU AAG 3′
- DNA- 3′ TTT TAC TCA TTC 5′
- The question is asking if the following pairing is accurate
- Each nucleotide is appropriately paired according to the A-U/T-A and C-G pairing rules, so these are complementary strands.
- 3′ TTT TAC TCA TTC 5′
- 5′ AAA AUG AGU AAG 3′. Which of the following DNA strands could have been the template for this RNA?
- Transcription has three stages: initiation, elongation, and termination. Match the following events with the correct stage of transcription.
| Initiation | Elongation | Termination | |
| RNA polymerase stops transcribing. | Yes | ||
| RNA polymerase binds to the promoter. | Yes | ||
| RNA polymerase adds complementary nucleotides to the 3′ end of the RNA strand. | yes |
- During initiation, RNA polymerase binds to a promoter on a DNA template strand. This tells the polymerase where to begin transcribing.
- Elongation is the stage when the RNA strand gets longer as new nucleotides are added by the polymerase.
- Termination is the process of ending transcription. It happens once the RNA polymerase transcribes a sequence of DNA known as a terminator.
- Which of the following is true regarding the process of transcription?
- Transcription occurs for individual genes.
- Transcription is controlled separately for each gene in your genome. Cells carefully ensure that just the genes whose products are needed at a particular moment are transcribed.
- Transcription occurs for individual genes.
- The following DNA strand is used as a template for transcription:
- 3′ CGTAAGCGGCT 5′. Which of the following RNA strands will be produced?
- 5′ GCAUUCGCCGA 3′
- The question is asking if the following pairing is accurate
- RNA – 5′ GCAUUCGCCGA 3′
- DNA – 3′ CGTAAGCGGCT 5′
- Each nucleotide is appropriately paired according to the A-U/T-A and C-G pairing rules, so these are complementary strands.
- 3′ CGTAAGCGGCT 5′. Which of the following RNA strands will be produced?
- When RNA transcript is first made in a eukaryote, it requires further processing to become messenger RNA (mRNA). What steps are needed to produce a mature mRNA?
- Introns are removed from the sequence.
- To produce a mature mRNA, introns are removed from the sequence. These sequences do not code for anything and must be removed in order for the RNA to encode the correct protein.
- Exons are joined together to form the mature mRNA.
- Exons are the segments of genes that code for proteins. Once the introns are removed, the exons are spliced together by the spliceosome to make the final, mature mRNA.
- Introns are removed from the sequence.
Transcription and RNA processing:
- The following diagram shows a double-stranded DNA molecule. Which of the following represents the correct mRNA molecule that will result from the transcription of the DNA molecule?


- The following diagram shows a double-stranded DNA molecule. Which of the following represents the correct mRNA molecule that will result from the transcription of the DNA molecule?


- Which of the following represents the correct mRNA molecule that will result from the transcription of the DNA molecule?


- Which of the following best represents how a single gene can encode more than one type of protein?

- Which of the diagrams best represents alternative splicing, and why?

- Diagram A, because it shows a single gene giving rise to two mRNAs, each of which is translated into a unique protein.
- Which diagram best represents how a single gene can encode more than one type of protein, and why?
- Diagram 2, because it shows the splicing together of exons to create more than one unique mRNA.

- Which of the following describes an event that occurs during Step 1 in the diagram?
- pre-mRNAs are produced by RNA polymerase using a template molecule.
- Step 1 represents transcription, or the production of pre-mRNAs from a DNA template. This step is carried out by RNA polymerase.
- pre-mRNAs are produced by RNA polymerase using a template molecule.
- Which of the following describes an event that occurs during Step 3 in the diagram?

- tRNAs are used to add amino acids to a growing polypeptide chain.
- Step 3 represents the translation of mRNA into protein. During translation, tRNAs are used to add specific amino acids to a growing polypeptide chain.
- Which of the following describes an event that occurs during Step 2 in the diagram?
- pre-mRNAs are modified to include only certain exons.
- Step 2 represents pre-mRNA processing. During this processing, pre-mRNAs undergo splicing so that only certain exons are included in the mature mRNA.
- pre-mRNAs are modified to include only certain exons.
- A group of scientists proposes that one of the mutations seen in patients with schizophrenia affects the alternative splicing of DRD2 pre-mRNAs in the brain. To test their claim, the scientists compare the sequence and abundance of mature DRD2 mRNAs in individuals with and without the mutation.Which of the following pieces of experimental evidence would best support the claim that alternative splicing of DRD2 pre-mRNAs is associated with the mutation described above?

- DRD2 mRNAs that include Exon 6 are more common in individuals with the mutation, while DRD2 mRNAs that exclude Exon 6 are more common in individuals without the mutation.
- This evidence shows that DRD2 pre-mRNAs can be alternatively spliced, with Exon 6 being either spliced out of or retained in mature mRNAs. Individuals with the mutation have a different common version of mature DRD2 mRNA compared to individuals without the mutation, suggesting that alternative splicing of DRD2 pre-mRNAs is occurring in individuals with the mutation.
- Which of the following pieces of experimental evidence would best support the claim that alternative splicing of NFE2L2 pre-mRNAs is associated with lung cancer in humans?
- NFE2L2 mRNAs without Exon 2 are more common in tumors, while those with Exon 2 are more common in normal tissue.
- This evidence shows that NFE2L2 pre-mRNAs are alternatively spliced, with Exon 2 being retained in some mRNAs and spliced out of others. A different version of mature NFE2L2 mRNA is more common in cancer tissue compared to normal tissue, suggesting that alternative splicing is associated with lung cancer in humans.
- NFE2L2 mRNAs without Exon 2 are more common in tumors, while those with Exon 2 are more common in normal tissue.

Cystic fibrosis, a disease that causes sticky mucus to build up in a person’s organs, is caused by mutations in the CFTR gene. The structure of the CFTR gene is shown in the following diagram, where rectangles represent exons and horizontal lines represent introns.
A group of scientists predicts that one of the mutations seen in cystic fibrosis patients causes alternative splicing of CFTR pre-mRNAs. To test their claim, the scientists compare the sequence and abundance of mature CFTR mRNAs in individuals with and without the mutation.
Which of the following pieces of experimental evidence would best support the claim that alternative splicing of CFTR pre-mRNAs is occurring in individuals with the mutation described above?

- CFTR mRNAs that exclude Exon 16 are more common in individuals with the mutation, while CFTR mRNAs that include Exon 16 are more common in individuals without the mutation.
- This evidence shows that CFTR pre-mRNAs can be alternatively spliced, with Exon 16 being either spliced out of or retained in mature mRNAs. Individuals with the mutation have a different common version of mature CFTR mRNA compared to individuals without the mutation, suggesting that alternative splicing of CFTR pre-mRNAs is occurring in individuals with the mutation.
Translation:
- What is the anticodon corresponding to the codon GUA
- CAU
- An anticodon is complementary to the codon. It follows the A-U and C/G base pairing rules: {GUA} – {CAU}
- CAU
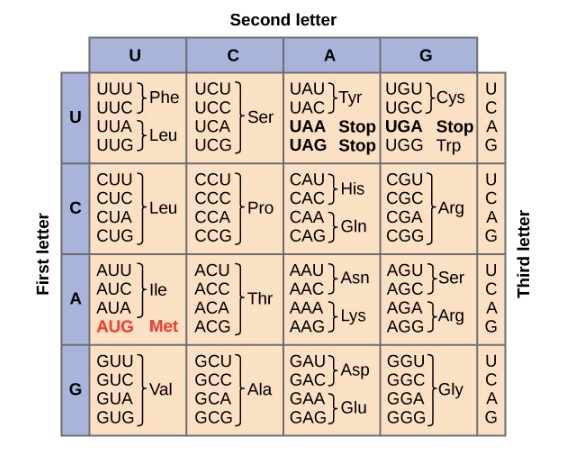
- A sequence of RNA is shown below:Using the codon chart provided, what is the sequence of amino acids that is produced when this gene is translated?
- Thr – Lys – Asp
- Since we are already given RNA, all we have to do is look at the chart to determine the appropriate amino acid. Remember that each group of three bases codes for one amino acid. Using the codon chart, we find that
- {ACG} codes for threonine (Thr)
- {AAA} codes for lysine (Lys)
- {GAU} codes for aspartic acid (Asp)
- Since we are already given RNA, all we have to do is look at the chart to determine the appropriate amino acid. Remember that each group of three bases codes for one amino acid. Using the codon chart, we find that
- Thr – Lys – Asp
- An RNA sequence is shown below.
- AUGCCGAAACGU
- Which of the following statements describes how the RNA sequence specifies the production of an amino acid chain?
- Each set of three bases codes for a single amino acid.
- The instructions for building a polypeptide are RNA nucleotides read in groups of three called codons.
- Each set of three bases codes for a single amino acid.
- Which of the following statements describes how the RNA sequence specifies the production of an amino acid chain?
- AUGCCGAAACGU
- A deletion mutation occurs, leaving 11 bases in nucleotide sequence. What is the maximum number of amino acids that could be coded for by this sequence?
- 3
- Every three bases codes for an amino acid. If we have 11÷3, that gives us three sets of three bases with two bases leftover. The remaining two bases will not be used to code for an amino acid, since three bases are needed.
- 3
Codons and mutations
- A deletion mutation occurs, leaving 14 nucleotides in a DNA sequence. What is the maximum number of amino acids that could be coded for by this sequence?
- 4
- Every three nucleotides (codon) codes for an amino acid. If we have 14÷3 that gives us four sets of three nucleotides with two left over.
- 4
- Below are two sequences of a segment of DNA.
- Normal sequence – TAA GCG CAT
- Mutated sequence – TAG CGC AT
- Deletion mutation – The mutated sequence shows the deletion of adenine (A) from the first codon.
- Below are two sequences of a segment of DNA.
- Normal sequence – CCG GTC TAG
- Mutated sequence – CCG GTC GTA G
- Which type of mutation has occurred?
- Insertion mutation – A guanine (G) nucleotide has been inserted between the second and third codons.
- Which type of mutation has occurred?
- Normal sequence – TAG GTC CAC
- Mutated sequence – TAG GTC CCC
- Which type of mutation has occurred?
- Substitution mutation
- The mutated sequence shows the substitution of cytosine (C) for adenine (A) in the third codon.
- Substitution mutation
- Which type of mutation has occurred?
- Mutated sequence – TAG GTC CCC
- Normal – GAC TAG CGA
- Mutated – GAC TTA GCG A
- Insertion mutation – A thymine (T) nucleotide has been inserted between the first and second codons.
- Mutated – GAC TTA GCG A
- Normal sequence – TGA CAC TAG
- Mutated sequence – TGA CCT AG
- Which type of mutation has occurred?
- Deletion mutation – The mutated sequence shows the deletion of adenine (A) from the second codon.
- Which type of mutation has occurred?
- Mutated sequence – TGA CCT AG
- Normal sequence – CAT TAC ACC
- Mutated sequence – CTT TAC ACC
- Which type of mutation has occurred?
- Substitution mutation – The mutated sequence shows the substitution of thymine (T) for adenine (A) in the first codon.
- Which type of mutation has occurred?
- Mutated sequence – CTT TAC ACC
- Normal sequence – TTA AAA GGA
- Mutated sequence – CTT AAA AGG A
- Which type of mutation has occurred?
- Insertion mutation – A cytosine (C) nucleotide has been inserted before the first codon.
- Which type of mutation has occurred?
- Mutated sequence – CTT AAA AGG A
- An insertion mutation occurs, leaving 31 nucleotides in a DNA sequence.What is the maximum number of codons that could be produced by this sequence?
- 10
- Every three nucleotides is a codon. Each codon codes for an amino acid. If we have 31÷3, that gives us 10 codons, with one nucleotide left over.
- 10
Gene regulation in bacteria:
- The lac operon is an inducible operon in E. coli that codes for genes involved in the breakdown of lactose. Which of the following statements is true regarding the lac operon?
- It regulates the production of proteins involved in the metabolism of lactose.
- The lac operon of E. coli contains genes involved in lactose metabolism. It’s expressed only when lactose is present and glucose is absent.
- It regulates the production of proteins involved in the metabolism of lactose.
- The trp operon of E. coli is a repressible operon responsible for producing enzymes involved in the synthesis of the amino acid tryptophan. Which of the following statements is true regarding the trp operon?
- When tryptophan is in short supply, the trp operon is expressed.
- The trp operon is expressed when tryptophan levels are low and repressed when they are high.
- When tryptophan is in short supply, the trp operon is expressed.
- Which of the following is true regarding gene regulation in prokaryotes such as bacteria?
- Prokaryotic gene expression can be inducible or repressible.
- Some operons are inducible, meaning that they can be turned on by the presence of a particular small molecule. Others are repressible, meaning that they are on by default but can be turned off.
- Prokaryotic genes are usually transcribed as a group and have a single promoter.
- Bacterial genes are often clustered in a chromosome and are transcribed from one promoter as a single unit. These gene clusters are called operons.
- Prokaryotic gene expression can be inducible or repressible.
- There are several components involved in bacterial operons. Which of the following correctly pairs the operon component with its function?
- A promoter is the site where RNA polymerase binds to the operon.
- Promotors are regulatory sequences that act as the binding site for RNA polymerase.
- A promoter is the site where RNA polymerase binds to the operon.
- Which of the following is true regarding gene regulation in prokaryotes such as bacteria?
- Prokaryotic gene expression can be inducible or repressible.
- Some operons are inducible, meaning that they can be turned on by the presence of a particular small molecule. Others are repressible, meaning that they are on by default but can be turned off.
- Prokaryotic genes are usually transcribed as a group and have a single promoter.
- Bacterial genes are often clustered in a chromosome and are transcribed from one promoter as a single unit. These gene clusters are called operons.
- Prokaryotic gene expression can be inducible or repressible.
- Transcription of genes by RNA polymerase requires energy, so a bacteria’s operon is usually not always turned on. In E. coli bacteria, the trp operon encodes genes for the synthesis of tryptophan, and the lac operon encodes genes for the digestion of lactose. Which of the following strains of E. coli is likely to use its energy most efficiently?
- A strain in which RNA polymerase is inhibited from transcribing the trp operon when tryptophan is present
- The trp operon is repressible, so the most energy-efficient scenario is to produce the enzymes to synthesize tryptophan when there is no tryptophan present (inactive repressor), and to be off (active repressor) when tryptophan levels are high.
- A strain in which RNA polymerase is inhibited from transcribing the trp operon when tryptophan is present
- The lac operon is an inducible operon in E. coli that codes for genes involved in the breakdown of lactose. Which of the following statements is true regarding the lac operon?
- It regulates the production of proteins involved in the metabolism of lactose.
- The lac operon of E. coli contains genes involved in lactose metabolism. It’s expressed only when lactose is present and glucose is absent.
- It regulates the production of proteins involved in the metabolism of lactose.

Recent Comments